Zemfira N. Karamysheva, Ph.D.
Associate Professor

Ph.D. Molecular Biology
Moscow State University
Curriculum Vitae
Department of Cell Biology and Biochemistry
Texas Tech University Health Sciences Center
3601 4th Street, Mail Stop 6540
Lubbock, TX 79430
Phone: (806) 743-3231
Fax: (806) 743-2990
zemfira.karamysheva@ttuhsc.edu
Research Interests
- Fundamental mechanisms of regulation of gene expression and its dysregulation in human diseases
- Translation control in protozoa and mammalian organisms
- Ribosome specialization
- Specialized ribosomes in Leishmania and their role in survival and adaptation of parasites during development and change of the host
- Drug resistance in Leishmania parasites
- RNA degradation pathways in protozoa and mammals
- mRNA translation regulation in health and disease
- Protein/mRNA quality control at the ribosome
Current Projects
Ribosome Specialization in Protozoa Parasites
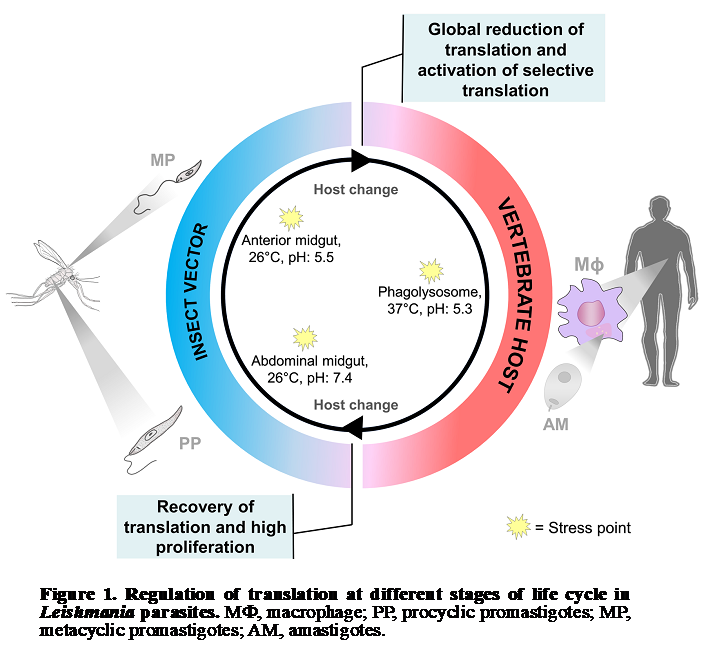 This project is focused on regulation of gene expression, translational control and
ribosome specialization in protozoa parasites. Specifically, we study the role of
ribosome specialization in protozoan parasite differentiation during transmission
from insect vector to mammalian host. Leishmania parasites alternate between promastigotes
living in the midgut of sandflies and amastigotes residing in the mammalian host (Fig.
1). The change between hosts involves dramatic temperature, pH, and nutritional stresses.
The adaptation to each host and parasite survival in harsh environment is achieved
through differential gene expression. Interestingly, Leishmania major lacks promoter-mediated
regulation of transcription and its gene expression is predominantly controlled at
the level of mRNA translation and stability. Our project is based on the hypothesis
that Leishmania ribosomes undergo change in their composition in order to promote
parasite survival and withstand environmental insults during transmission from insect
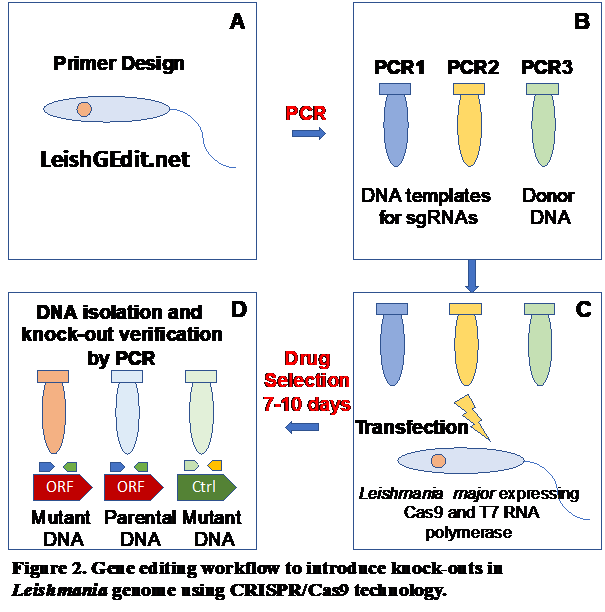
vector to mammalian host. We developed an integrative approach consisting of polysome
profiling followed by deep RNA sequencing of translatomes, proteomic analysis of ribosomes
and CRISPR-Cas9 gene editing knock-out screens for gene function investigation under
normal and stress conditions (Fig. 2).
This project is focused on regulation of gene expression, translational control and
ribosome specialization in protozoa parasites. Specifically, we study the role of
ribosome specialization in protozoan parasite differentiation during transmission
from insect vector to mammalian host. Leishmania parasites alternate between promastigotes
living in the midgut of sandflies and amastigotes residing in the mammalian host (Fig.
1). The change between hosts involves dramatic temperature, pH, and nutritional stresses.
The adaptation to each host and parasite survival in harsh environment is achieved
through differential gene expression. Interestingly, Leishmania major lacks promoter-mediated
regulation of transcription and its gene expression is predominantly controlled at
the level of mRNA translation and stability. Our project is based on the hypothesis
that Leishmania ribosomes undergo change in their composition in order to promote
parasite survival and withstand environmental insults during transmission from insect
vector to mammalian host. We developed an integrative approach consisting of polysome
profiling followed by deep RNA sequencing of translatomes, proteomic analysis of ribosomes
and CRISPR-Cas9 gene editing knock-out screens for gene function investigation under
normal and stress conditions (Fig. 2).
Our data reveal a dramatic change in the ribosome composition supporting our hypothesis and the feasibility of the approach. Currently, we examine the role of specialized ribosome components in parasite survival and differentiation using polysome profiling and CRISPR/Cas9 gene knock-out screen.
Translational reprogramming as a driver of antimony drug resistance in Leishmania parasites
The second project is focused on the molecular mechanisms of drug resistance in Leishmania parasites. Specifically, we explore the role of translational reprogramming in the development of drug resistance. Current studies on drug resistance in Leishmania are mostly limited to the research at genomic level. However, changes at genomic level cannot explain all possible mechanisms of resistance and treatment failures without genetic-based resistance are very widespread. Our data demonstrate that Leishmania parasites undergo a dramatic reprogramming of mRNA translation during development of drug resistance. As a consequence, it leads not only to changes in proteome but also to lipidome and metabolome remodeling thus contributing to drug resistance. We propose a novel concept just published in Nature Communications (https://www.nature.com/articles/s41467-023-38221-1) that establishes translational control as a major driver of antimony drug resistance in Leishmania. In future directions gene editing knock-out screen and drug assays will be performed to validate targets involved in drug resistance.
Translational control and stress responses
Humans of all ages are exposed to a plethora of dangerous chemicals and environmental stressors present in contaminated water, air, soil and food. These environmental exposures can induce severe health effects even in modest concentrations. The evidence is emerging now that common contaminants such as heavy metals, per- and polyfluorinated alkyl substances (PFAS), nano plastic, herbicides and pesticides are associated with cancer, fertility issues, birth defects, cardiovascular and neurodegenerative diseases and accelerated aging. However, the mechanistic studies on molecular mechanisms of the cellular and molecular responses to environmental stressors leading to clinically relevant disease outcomes are lacking. In our lab we study the impact of environmental insults on the molecular pathways in the cell and how it contributes to human diseases.
Weblink to the Lab
Visit our lab page at https://karamyshevaslab.wixsite.com/karamysheva-s-lab
Graduate students inspired by science are welcome to the lab!
Please contact Dr. Zemfira Karamysheva if you have an interest to join the lab: zemfira.karamysheva@ttuhsc.edu
Selected Publications
Full publications list at NCBI Bibliography: https://www.ncbi.nlm.nih.gov/myncbi/1ZUlk-i9v8hAh/bibliography/public/
- Gutierrez Guarnizo, S. A., Kellogg, M. K., Miller, S. C., Tikhonova, E. B., Karamysheva, Z. N. Karamyshev, A. L. (2023) Pathogenic Signal Peptide Variants in the Human Genome. NAR Genomics and Bioinformatics, 5(4). https://doi.org/10.1093/nargab/lqad093
- Tarannum, A., Rodríguez-Almonacid, C.C., Salazar-Bravo, J., Karamysheva, Z.N.* Molecular Mechanisms of Persistence in Protozoan Parasites. Microorganisms. Published September 7, 2023, 11, 2248. https://doi.org/10.3390/microorganisms11092248
- Karamysheva, Z. N.*, Karamyshev, A. L.* (2023) Aberrant Protein Targeting Activates Quality Control on the Ribosome. Frontiers in Cell and Developmental Biology. Volume 11. Published June 6, 2023. doi: 10.3389/fcell.2023.1198184
- Gutierrez Guarnizo S. A., Tikhonova E. B., Karamyshev A. L.*, Muskus C.E.*, Karamysheva Z. N.* Translational reprogramming as a driver of antimony-drug resistance in Leishmania.
Nature Communications 2023 May 5;14(1):2605. doi: 10.1038/s41467-023-38221-1. PMID: 37147291; PMCID: PMC10163012.
The article has been chosen by the Senior Editor Madlen Luckner for the Editors’ Highlights in Microbiology and Infectious Diseases: https://www.nature.com/ncomms/editorshighlights
The article has been featured in Texas Tech Today on June 1, 2023. Finding Possible Solutions Through Working With Parasites by Doug Hensley: https://today.ttu.edu/posts/2023/06/Stories/Finding-Possible-Solutions-Through-Working-With-Parasites TTUHSC Daily Dose by Mark Hendricks. TTUHSC-TTU Research Collaboration Leads to Possible Drug Targets for Leishmaniasis (by Mark Hendricks). https://dailydose.ttuhsc.edu/2023/june/research-collaboration-Leishmaniasis.aspx
EurekAlert! AAAS TTUHSC-TTU research collaboration leads to possible drug targets for Leishmaniasis: https://www.eurekalert.org/news-releases/993169 - Rodríguez-Almonacid CC, Kellogg MK, Karamyshev AL, Karamysheva Z.N.* Ribosome Specialization in Protozoa Parasites. Int J Mol Sci. 2023 Apr 19;24(8):7484. doi: 10.3390/ijms24087484. PMID: 37108644; PMCID: PMC10138883.
- Miller S. C, MacDonald C. C, Kellogg M. K, Karamysheva Z.N., Karamyshev A. L. Specialized Ribosomes in Health and Disease. Int J Mol Sci. 2023 Mar 28;24(7):6334. doi: 10.3390/ijms24076334. PMID: 37047306; PMCID: PMC10093926.
- Tikhonova, E. B., Gutierrez Guarnizo, S. A., Kellogg, M. K., Karamyshev, A., Dozmorov, I. M., Karamysheva, Z. N., Karamyshev, A. L. (2022) Defective Human SRP Induces Protein Quality Control and Triggers Stress Response. Journal of Molecular Biology, vol. 434, Issue 22. https://doi.org/10.1016/j.jmb.2022.167832
- Karamysheva, Z. N.*, Moitra S., Perez A., Mukherjee S., Tikhonova E.B., Karamyshev A.L., Zhang K.* (2021). "Unexpected Role of Sterol Synthesis in RNA Stability and Translation in Leishmania." Biomedicines, 9(6): 696; https://doi.org/10.3390/biomedicines9060696
- Gutierrez Guarnizo, S. A., Karamysheva, Z. N., Elkin G., Muscus, C. Metabolite biomarkers of Leishmania antimony resistance, Cells, (2021), 10(5), 1063; https://doi.org/10.3390/cells10051063.
- Gutierrez Guarnizo, S. A., Tikhonova E.B., Zabet-Moghaddam M., Zhang K., Muscus, C., Karamyshev, A. L.*, Karamysheva, Z. N.* Drug-induced lipid remodeling in Leishmania parasites, Microorganisms, (2021), 9(4), 790; https://doi.org/10.3390/microorganisms9040790.
- Karamysheva, Z. N.*, Gutierrez Guarnizo, S. A., Karamyshev, A. L.* (2020) Regulation of Translation in the Protozoan Parasite Leishmania, Review, Int. J. Mol. Sci., 21, 2981; doi:10.3390/ijms21082981.
- Karamyshev A. L.*, Tikhonova, E. B., Karamysheva, Z. N.* (2020) Translational Control of Secretory Proteins in Health and Disease. Review, Int. J. Mol. Sci., 21, 2538; doi:10.3390/ijms21072538.
- Karamysheva, Z. N.*, Tikhonova, E. B., Karamyshev, A. L.* (2019) Granulin in Frontotemporal Lobar Degeneration: Molecular Mechanisms of the Disease. Review, Frontiers in Neuroscience, doi: 10.3389/fnins.2019.00395. PMID: 31105517 PMCID: PMC6494926.
- Karamyshev, A. L.*, Karamysheva, Z. N.* Lost in Translation: Ribosome-Associated mRNA and Protein Quality Controls. Review, Front. Genet., 2018, Oct 4;9:431. PMID:30337940 PMCID: PMC6180196.
- Karamysheva, Z. N., Tikhonova, E. B., Grozdanov, P. N., Huffman J. C., Baca, K. R., Karamyshev, A, Denison R. B., MacDonald, C. C., Zhang, K., Karamyshev, A. L. (2018) Polysome Profiling in Leishmania, Human Cells and Mouse Testis. Journal of Visualized Experiments (JoVE), Apr 8; (134) doi: 10.3791/57600.
- Diaz-Martinez L.A., Karamysheva Z. N., Warrington R., Li B., Wei S., Xie X.J., Roth M.G. (2014) Genome-wide siRNA screen reveals coupling between mitotic apoptosis and adaptation. EMBO J., 33(17), 1960-1976.
- Karamyshev A.L., Patrick, A. E.*, Karamysheva, Z. N.*, Griesemer D.S., Hudson H., Tjon-Kon-Sang S., Nilsson I., Otto H., Liu Q., Rospert
S., von Heijne G., Johnson A.E., Thomas P.T. (2014) Inefficient SRP Interaction with
a Nascent Chain Recruits Ago2 and Triggers Specific mRNA Degradation. Cell, 156(1-2), 146-157.
Featured in TIBS: Popp, M. W.-L. and Maquat L. E. (2014) Defective secretory-protein mRNAs take the RAPP, Trends in Biochemical Sciences, Vol. 39, No. 4, p. 154-156.
Featured in Nature Reviews Genetics: Research highlights: Novel mRNA quality control mechanism. Nature Reviews Genetics (2014) vol. 15 (3), p. 144.
The article was selected by Faculty of 1000. - Karamysheva Z., Wang L., Shrode T., Bednenko J., Hurley L. A., Shippen D. E. (2003) Developmentally
programmed gene elimination in Euplotes crassus facilitates a switch in the telomerase
catalytic subunit. Cell, 113:565-576.
Preview on this paper has been published in Cell (2003) 113:552-554 by Cristofari G, Lingner J. Fingering the ends: how to make new telomeres.
Featured in Nature Reviews Molecular Cell Biology: Telomeres (2003) 4, p. 509. https://www.nature.com/articles/nrm1163 - Karamysheva Z. N, Karamyshev A. L., Ito K., Yokogawa T., Nishikawa K., Nakamura Y., Matsufuji S. (2003)
Antizyme frameshifting as a functional probe of eukaryotic translational termination.
Nucleic Acids Res., 31:5949-5956. Impact factor 19.16.
This paper was selected as technical advance by Faculty 1000. - Ray S., Karamysheva Z., Wang L., Shippen D. E., Price C. M. (2002) Interactions between telomerase and primase physically link the telomere and chromosome replication machinery. Mol. Cell. Biol., 22:5859-5868.
